Figure 1. The appearance of Human kinase classes
throughout eukaryotic evolution
The sponge, Amphimedon queenslandica, sits at a crucial junction in evolutionary history. As one of the simplest and earliest branching metazoans, it offers key insights into the explosion of kinase complexity around the emergence of multicellualar animals. By comparing this kinome to other early branching metazoans, and to their closest unicellular relatives (such as the choanoflagellate Monosiga brevicollis), we are able to track the emergence of well established metazoan kinase classes and kinase signalling pathways. Our analysis of the sponge kinome is part of the sponge genome paper (below), and forms a core part of our work to understand the evolution of the metazoan kinome.
The genome of the haplosclerid demosponge Amphimedon queenslandica and the evolution of animal complexity.
Srivastava, M, Simakov, O, Chapman, J, Mitros, T, Hellsten, U, Putnam, NH, Fahey, B, Gauthier, M, Larroux, C, Richards, G, Stanke, M, Adamska, M, Darling, A, Dacre, M, Degnan, S, Zhai, Y, Adamski, M, Calcino, A, Goodstein, DM, Shengqiang Shu, S, Woodcroft, B, Martindale, M, Leys, S, Manning, G, Degnan, B, Rokhsar, DS.
Nature (2010) 466:720-726 (Medline, PDF)
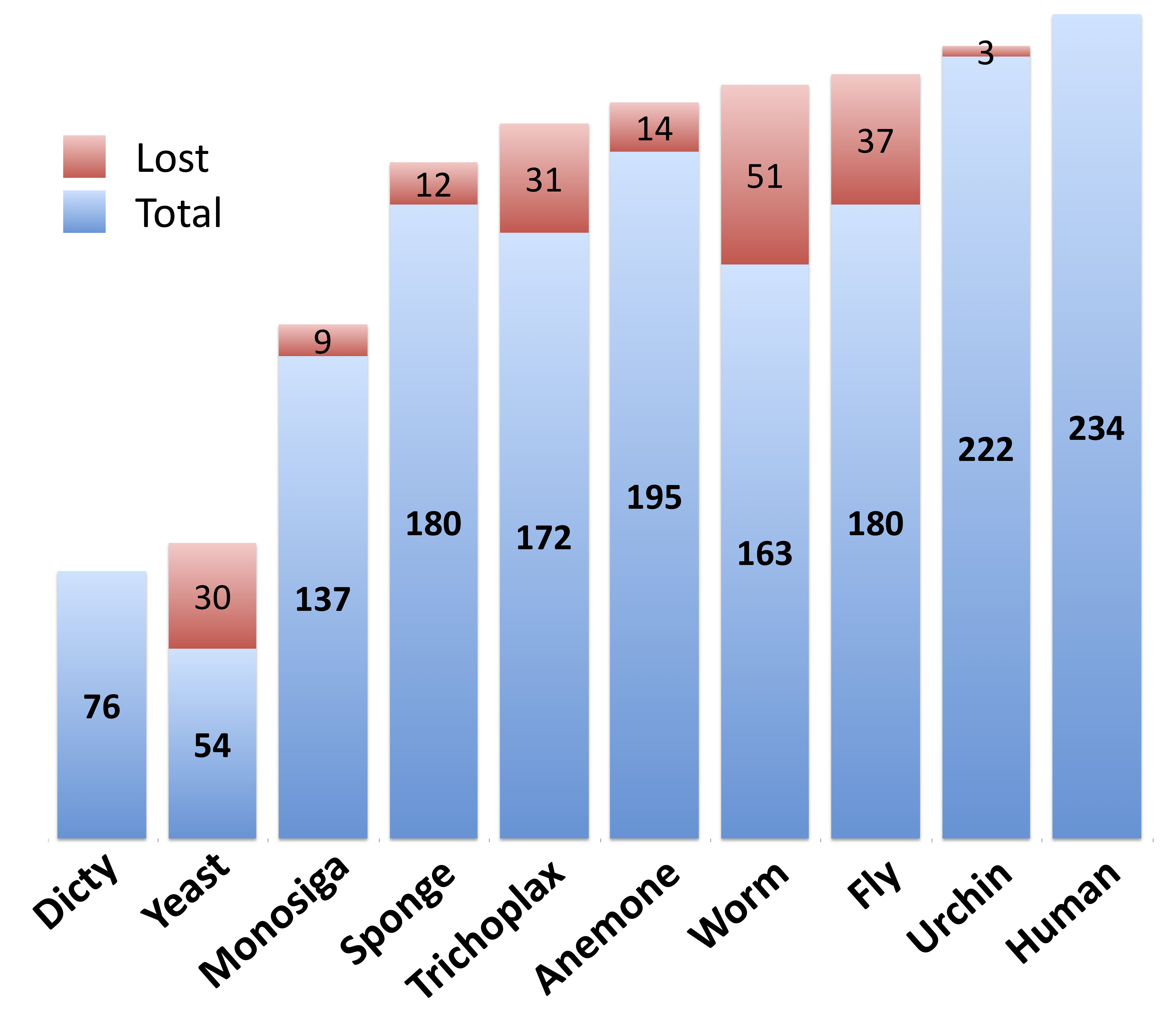
We find that the majority of of kinase complexity appears very early in the animal lineage: while just 23% of human kinase classes are found in yeast, 74% have orthologs in Amphimedon (and 56% have orthologs in Monosiga). This number rises to 83% in sea anemone (Nematostella vectensis). Interestingly, we actually see a large loss of Human kinase classes in the model organisms Drosophila and C. elegans - with just 77% and 70% respectively (figure 1).

More than half of the Amphimedon kinome is found in 11 expanded classes of the tyrosine kinase (TK) and tyrosine kinase like (TKL) groups. Of the tyrosine kinases, a core of ~58 are recognizable orthologs of kinases present in more complex metazoa, the remaining ~173 are apparently unique to Amphimedon and have expanded to suit the needs of its environment. We see a broad range of kinases with diverse extra-cellular domains, the functions of which we can only guess at.
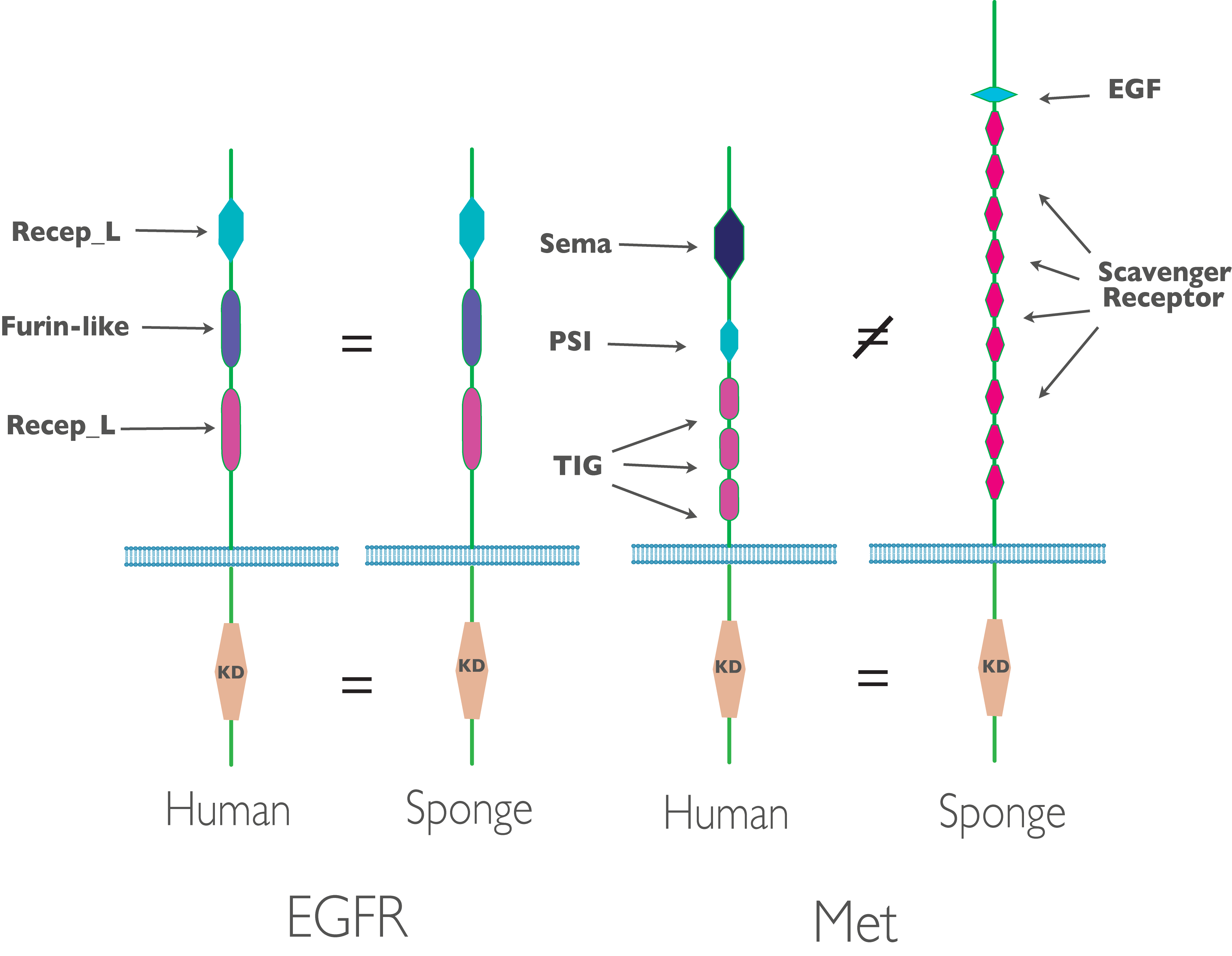
In the receptor tyrosine kinases (RTKs) that are recognizable orthologs to known RTKs in human, we see interesting patters of conservation: in some cases, such as EGFR, we see fully developed human-like kinases with conservation of both the cytoplasmic kinase domain and the extra-cellular domains. In other (e.g. Met), we see conservation of just the cytoplasmic kinase domain, with little or no similarity in extra-cellular domain. This makes sense considering human receptor kinases point inwards - into the intercellular space. The majority of sponge cells point outwards - into the sea water the sponge filters for nutrition (figure 2).
The kinome is still incomplete: Many predicted kinases were fragmentary and mapped to internal gaps or contig ends. Other apparent fragments map within gapless regions, but are difficult to assemble due to the lack of close homologs, or possible assembly artefacts. We used homology to improve many gene predictions, but have not attemped to optimize using sponge paralogs, so many kinase sequences may still be incorrect or incomplete. 23 fragments with greater than 95% amino acid identity to other predictions that originated from short contigs were removed as probable assembly errors.
All sponge kinome sequences and classification are now available through our KinBase database, along with domain structures and alignments.
The kinome analysis was lead by Michael Dacre and Gerard Manning, with computational help from Yufeng Zhai and support from other members of the Manning lab.